New method pinpoints individual signaling pathways in cell
A new technique that can uncover the contribution of a single molecular signaling pathway among the hundreds of thousands of other reactions taking place in a cell at any given time has been developed by researchers at the University of Illinois at Chicago College of Medicine.
Their method, published in the journal Nature Chemical Biology, will let researchers study the influence of individual signaling pathways on cellular behavior and could illuminate new targets for drugs that would work with unprecedented specificity.
To determine the role of a specific protein, researchers typically delete its gene and see what happens to the cell. But if that protein is involved in multiple cellular processes, it can be very hard to determine its role in any one function.
“A single protein is like the conductor of an orchestra, directing multiple musicians that together create a complex piece of music,” says Andrei Karginov, assistant professor in pharmacology, the study’s first author. “But it’s hard to accurately detect the contribution of a single violin in the orchestra.” Deleting a protein entirely, he said, is the equivalent of removing the conductor and silencing the whole orchestra.
“Now,” Karginov said, “we can preserve and control the function of the protein conductor, so to speak, and tease out the relationship between that protein and one of the many molecules it interacts with and observe how that specific cellular pathway influences cellular behavior.”
Karginov and his colleagues were focused on the role of an enzyme called Src, which catalyzes multiple reactions, including some involved in cell movement.
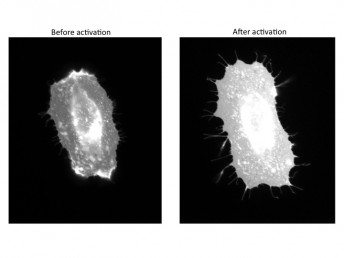
They developed a novel technique that allowed them to study the effect of activating just two of the myriad molecular pathways catalyzed by Src. They knew that these two pathways were important for cell movement, but didn’t know the exact contribution of each pathway to the process.
Karginov found that when one of the pathways was activated, the cell was able to change shape to form arm-like protrusions – a crucial first step in cell movement. When the other pathway was stimulated, the cell dramatically rearranged the adhesion points that help to pull it across the surface. The exact role of Src in these two pathways was previously unknown.
“Src also plays a role in the uncontrolled growth of cancer cells,” Karginov said. “With this new technique, we may be able to identify the exact Src pathways involved in this kind of growth and develop drugs to interfere with only those pathways without disrupting the protein’s ability to perform all its other functions.”
But the technique the researchers developed for isolating and studying individual cellular molecular pathways “may be the more exciting result of our research,” Karginov said. “It is an important new tool that can be used by other investigators to isolate individual cellular process and can also help identify new targets for drugs that would act with much greater specificity.”
Denis Tsygankov, Matthew Berginski, Pei-Hsuan Chu, Evan Trudeau, Jason Yi, Shawn Gomez, Timothy Elston and Klaus Hahn from the departments of pharmacology, biomedical engineering and the Lineberger Cancer Center at University of North Carolina at Chapel Hill are co-authors on the paper.
The research was funded by grants R21CA159179, GM102924 and GM094663 from the National Institutes of Health.