New ribosome-targeting antibiotic acts against drug-resistant bacteria
A new study published in Nature reports on a new antibiotic that binds to the ribosome of bacterial cells and stops drug-resistant pathogens from making mice sick.
Co-authored by researchers from the University of Illinois Chicago, the study not only shows the potential of the drug — called iboxamycin — to one day help humans who are ill because of antibiotic-resistant bacteria, but also identifies how the drug overcomes the most widespread mechanism of resistance to this class of antibacterials.
The drug — a synthetic oxepanoprolinamide, which is a novel class of antibacterial drugs — was developed and tested in animals by study co-authors from Harvard University.
The Nature study, “A synthetic antibiotic class overcoming bacterial multidrug resistance,” reports that iboxamycin was powerfully effective at fighting both gram-negative and gram-positive drug-resistant bacteria in mouse models.
The study also reports on the UIC discovery: the molecular mechanism that allows this drug to overcome resistance, which is important information the scientific community can use to make more informed decisions when searching for and developing new antibiotics and designing studies to test them.
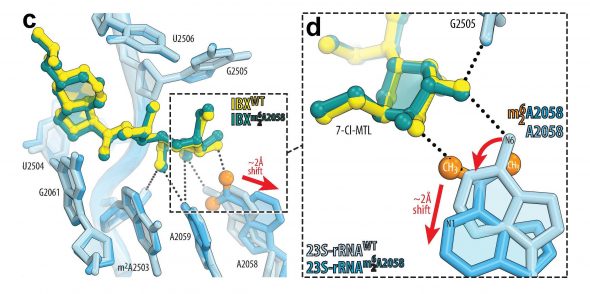
“It was exciting to see this agent bound in the structure of a drug-resistant ribosome, and it was quite surprising that the drug binds in the same exact way as to the regular ribosome but causes significant structural re-arrangements that has never been observed before or could be predicted from the existing data,” said study co-corresponding author Yury Polikanov, UIC associate professor of biological sciences at the College of Liberal Arts and Sciences and associate professor of pharmaceutical sciences at the College of Pharmacy.
In previous research, Polikanov developed a unique approach for visualizing ribosomes that are resistant to conventional antibiotics. Through this process, he and Alexander Mankin, UIC professor of pharmaceutical sciences, reported in a January Nature Chemical Biology paper that methylation — a process of changing the chemical make-up of a nucleotide in the ribosomal drug binding site — results in the inability of such ribosomes to bind some clinically important antibiotics rendering such ribosomes drug-resistant.
To understand how and why iboxamycin manages to overcome resistance, Polikanov and UIC graduate student Egor Syroegin, study co-first author, applied their unique visualization techniques. The UIC researchers co-crystallized bacterial ribosomes with the drug and froze the crystals. Then, they used a powerful X-ray at the Advanced Photon Source facility at Argonne National Laboratory to determine the molecule’s diffraction pattern — the places where the radiation bounced off the atoms in the crystals. This pattern was used to calculate electron density maps of the ribosome-drug complex and visualize their interactions.

“The whole process of atomic structure determination using X-rays is like a 3D puzzle,” Polikanov said. “We identified the open place in the ribosome puzzle, and we just needed to find the drug piece that fit. Once we did that, we could see where and how the drug was bound to the ribosome.”
When the UIC researchers looked at their visualization of the Harvard drug interacting with the drug-resistant ribosome, they found something unexpected.
“We noticed that the most common resistance mechanism via methylation of the ribosome does not work against this new drug because it still binds to the methylated ribosome and evades this type of resistance. We’ve also noticed that iboxamycin actually caused the methylated nucleotide at the heart of the ribosome and right at the drug binding site to move out of the drug’s way so that both the drug and the methylated nucleotide in the drug-resistant ribosome can co-exist — this was totally unexpected and unprecedented,” Polikanov said.
“This shows us Mother Nature is much smarter than we are. There is certainly a place for rational drug design, but we can’t forget the importance of simple trial and error,” he said. “Neither iboxamycin’s spectrum of activity, nor its potency in resistant strains, could have been predicted from prior knowledge.”
Additional UIC co-authors of the study include Mankin and Dorota Klepacki.
Andrew Myers, professor of chemistry and chemical biology, led the Harvard research team, which included study co-authors Matthew Mitcheltree, Amarnath Pisipati, Katherine Silvestre, Jeremy Mason, Daniel Terwilliger, Giambattista Testolin, Aditya Pote, Kelvin Wu, Richard Porter Ladley and Kelly Chatman.
The research at UIC was supported by state of Illinois startup funds and grants from the National Institutes of Health (R21AI137584, R01GM132302, R35GM127134). Additional funding for the research and disclosures are noted in the paper.
Categories
Health Sciences Colleges, Research
Topics
antibiotic resistance, antibiotics, ribosome, x-ray crystallography
